
The Effect of Exposure to Ethanol onTranscriptional Levels of Sonic Hedgehog in 2-Day Old Chicks
Jason Huska, Ben Myers, Sarah Albanese, Samantha Shenberger
Millersville University
Fetal alcohol syndrome is a condition that results when women drink excessive amounts of alcohol during their pregnancy, resulting in detrimental effects on the fetus. A child born with fetal alcohol syndrome can display complications such as poor growth and muscle tone, delayed cognitive development, heart defects, and structural malformations of the face. The child may also show significant functional problems in thought, speech, movement, or social skills as they age (Kilburn et al, 2006). Many of the symptoms of fetal alcohol syndrome may be attributed to changes in the cytoplasmic levels of certain transcription factors brought on by the ethanol circulating in the blood (Loucks and Ahlgren, 2009). We believe that changes in the transcription factor Sonic Hedgehog (SHH) may have adverse effects for the developing embryo.
In normal embryonic development, the sonic hedgehog pathway directs proper growth and development of neural tissue and facial features (Marti et al, 1995). A child born with fetal alcohol syndrome displays classic facial deformities as well as abnormalities in tissues deriving from the neural crest. This suggests that the SHH pathway may be inhibited or negatively affected by ethanol. In addition, previous experiments have shown that exogenous SHH administration negates the effects of ethanol on a developing embryo, saving it from deformities (Ahlgren et al, 2002). In this experiment, we will simulate fetal alcohol syndrome in chick embryos by exposing them to various concentrations of ethanol at 2 days post fertilization and study the effects this has on the development of the facial mesenchyme cells. The cytoplasmic levels of sonic hedgehog (SHH) will be analyzed to determine if the symptoms of fetal alcohol syndrome can be correlated to changes in the rate of SHH transcription. We predict that by exposing 2-day old chick embryos to ethanol, we will observe a down-regulation of SHH. To prove this, total RNA will be isolated from the affected tissues and amplified via RT-PCR using primers for SHH.
Methods
Ethanol Exposure
With beak-tipped forceps, a hole (2-5 mm in diameter) was made above the air space of 28, 2-day old chick embryos making sure not to puncture the yolk. Using a 25 gage needle and syringe, 250 μl of an ethanol solution (0%, 5%, 10%, or 20%) was injected into the yolk of the 28 embryos (n=7 for each group). Scotch tape was placed over the hole and the embryos were incubated at 37oC overnight.
Detection of Cell Death
Following the 24 hour incubation, the embryos were removed from the eggs and placed into Petri dishes of Howard RingerÕs solution. The embryos were then cleaned, removed from the amnion, and visualized under dissecting microscopes for any physical deformities. Embryos that were exposed to a particular ethanol solution were stained together with 1/10 dilute Trypan Blue to observe any cell death. Each embryo was then washed in Howard RingerÕs solution and digital photographs were taken for documentation.
Isolation of Chick Embryo Total RNA
The heads of the embryos were removed and each was placed into a 1.7 ml microfuge tube; all 7 from each group together in one. To each, 1 ml of guanidine isothiocyanate (TRIZOL; Sigma, St. Louis, MO) was added. The tissue was homogenized by aspiration and incubated at room temperature for 5 minutes. Following the incubation, 1 ml of each homogenate was transferred to clean microfuge tubes and 200 μl of chloroform was added to each. Each sample was mixed vigorously, incubated at room temperature for 3 minutes, and centrifuged at 11,500 rpm for 15 minutes. The aqueous layer from each was removed and transferred to a clean microfuge tube and 500 μl of isopropanol were added to pellet the RNA. Samples were mixed by inversion and incubated at room temperature for 10 minutes. This was followed by a 10 minute spin at 11,500 rpm and the supernatant was decanted. Each pellet was washed with 1 ml of 75% ethanol, spun at 9,500 rpm for 5 minutes, and dried. Each was resuspended in 40 μl of sterile ddH2O containing 2.5 U of RNA Guard and the total amount of RNA was quantified using a Nanodrop spectrophotometer.
Amplification of Sonic Hedgehog mRNA by RT-PCR
Into a 200 μl thin-wall PCR tube, 5 μl of total RNA were added and brought to 11 μl using ddH2O. Oligo (dT)12-18 was added at a final concentration of 0.5 μg/μl and the contents were incubated in a 65oC water bath for 5 minutes and then chilled on ice. dNTPÕs were added to final concentrations of 0.5 mM, first strand buffer to 1X, and DTT to 0.01M. RNaseOUT Recombinant Ribonuclease Inhibitor was added at 40U/reaction and the samples were incubated at 42oC for 2 minutes. SuperScript II RNase H Reverse Transcriptase was added at 200 U/reaction and mixed by aspiration. The reaction was incubated at 42oC for 50 minutes to generate cDNA. The enzyme was then inactivated by heating at 70oC for 15 minutes.
A master mix was made for six, 45 μl PCR reactions, each with a final PCR buffer concentration of 1X, 0.5 mM dNTPÕs, and 2.5 U of Fast Taq DNA polymerase. This mater mix was split into two equal volumes; added into one was primer mix for SHH cDNA (forward and reverse primer at 0.5 μM) and into the other primer mix for GADPH was added (forward and reverse primer at 0.5 μM). A 45 μl aliquot was removed from each and 5 μl of ddH2O were added. The remaining master mixes were split into 4, 45 μl reactions, with 2 reactions containing primers for GADPH and 2 other containing primers for SHH. cDNA from the 0% and 5% ethanol exposed embryos were added to each of one of the different primer reactions to a total volume of 50 μl. Each sample was denatured at 94oC for 2 minutes followed by 30 cycles of 94oC for 30 seconds, 60oC for 1 minute, and 72oC for 1 minute. The final cycle consisted of 72oC for 5 minutes and the sample were stored at 4oC until further use.
Detection of PCR Products by Agarose Gel Electrophoresis
A 1.5% agarose gel was prepared by dissolving agarose powder in 1X TBE and 10 ml were pipetted onto a glass plate. A comb was inserted into both and the gels were allowed to solidify. Samples were prepared by adding 1 μl of loading dye to 5 μl of each PCR reaction and loaded into the gel. Samples were size-separated by electrophoresis alongside 4 μl of HindIII digested λ phage DNA marker at 100 volts until the marker migrated half-way through the gel matrix. The gel was stained in 1 μg/ml ethidium bromide for 10 minutes and destained in 1X TBE for 5 minutes. Bands were visualized using UV light and digital photographs were taken for documentation.
Results
Four groups of 7, 2-day old chick embryos were injected with 250 μl of an ethanol solution; 0%, 5%, 10%, or 20%. After incubating for 24 hours at 37oC, the embryos were removed from the egg an analyzed for any physical deformities. All embryos injected with the 0% solution were properly developed and remained viable (Figure 1A). When observed with a dissecting microscope, the cranial features appeared normal and the heart was working properly. The embryos that were injected with the 5% ethanol solution displayed abnormal development that ranged in severity. Only 2 of the 7 appeared normal, however, when observed under the microscope, the heart had a weaker contraction as compared to the control. Three other embryos displayed moderate abnormality in the facial region and the midbrain was considerably smaller. The final two embryos from this group exhibited severe abnormalities as compared to others in their group. In these two specimens, the fore, mid, and hindbrain were extremely smaller than in other embryos and the heart was severely malformed and had a very low heart rate. Also, the tail curved outwards as opposed to the normal inward curve (Figure 1B). All embryos that were exposed the 10% and 20% ethanol solutions did not progress past the primitive streak stage and th majority only appeared as scattered bunches of cells.
Only the embryos injected with the 0% and 5% ethanol solutions were used to proceed with experimentation. Each embryo was stained with 1/10 trypan blue to detect cell death. All the embryos exposed to 5% ethanol displayed higher percentages of stained cells, most predominantly in the head regions. Those that had more severe abnormalities displayed greater amounts of cell death. In the embryos exposed to the 0% solution, minimal stained cells were observed in the facial region, however, cells did stain in the area near the limb buds.
Once cell death was confirmed, total RNA was isolated from the head region and used as a template for cDNA synthesis. The cDNA specific for SHH was amplified and size-separated by gel electrophoresis. Bands representing SHH cDNA from the embryos exposed to 5% ethanol were less intense when compared to the control, signifying fewer SHH cDNA molecules in this sample (Figure 2).
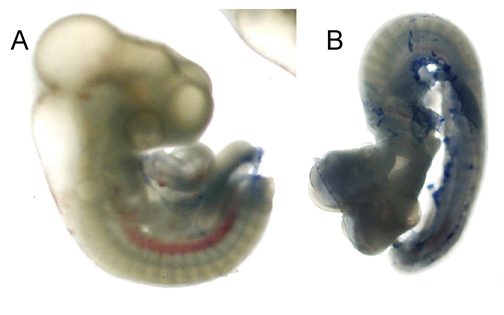
Figure 1 Trypan blue stained chick embryos. 2-day old chick embryos were injected with a 0% (A) and 5% (B) ethanol solution and incubated at 37oC overnight. The embryos were extracted from the egg, extra-embryonic membranes were removed, and embryos were stained with Trypan blue to observe any cell death.
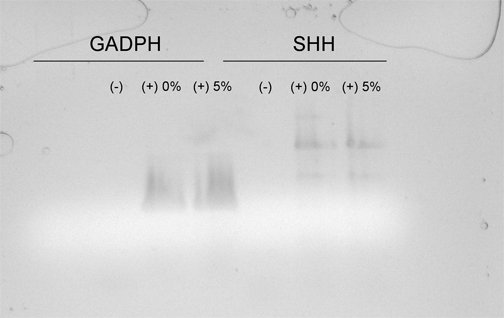
Figure 2 Amplification of SHH cDNA from 2-day old chick embryos exposed to either a 0% or 5% ethanol solution by RT-PCR. Total RNA was isolated from the cranial region of chick embryos and amplified using primers for SHH and GADPH cDNA. Lanes 1-3 contain primers for GADPH and lanes 4-6 contain primers for SHH. Lane 1 and 4 exclude cDNA, lanes 2 and 5 contain cDNA from embryos injected with 0% ethanol and lanes 3 and 3 containing cDNA from embryos injected with 5% ethanol.
Discussion
We have shown a decrease in the transcriptional levels of sonic hedgehog in 2-day old chick embryos after injection with a 5% ethanol solution. Changes in the cytoplasmic levels of SHH may be attributed the symptoms associated with fetal alcohol syndrome. The SHH signaling pathway is important in the development of the midline structures of the midbrain, spinal cord, and thalamus (Marti et al, 1995). If changes in the biochemical events occur within this pathway, the neural structures of the cranial region will not form properly. Excessive alcohol consumption during pregnancy is known to cause deformities in these structures, and our results have shown similar malformations in the exposed embryos. With these two sets of results in mind it is suggestive that SHH pathway is inhibited during development. When embryos were exposed to the 5% ethanol, we observed less cDNA for SHH, correlating to less mRNA in the cytoplasm available for translation. This suggests that ethanol may have negative effects on SHH at the transcriptional level. If less SHH is produced, cells may not be signaled to differentiate properly, leading to the symptoms associated with the disease (Loucks and Ahlgren, 2009). Furthermore, ethanol may activate the apoptotic pathway as indicated by cells stained by Trypan blue. As a result, larger percentages of the developing embryo may be undergoing cell death as compared to typical development. Further research will be needed to completely define the mechanism ethanol uses to change the transcriptional rate of SHH as well as determine potential apoptotic up-regulation.
References:
Ahlgren, S., Thakur, V., and Bronner-Fraser, M. (2002). Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc Natl Acad Sci U S A. 99: 10476-10481
Kilburn, B., Chiang, P., Wang, J., Flentke, G. Smith, S., and Armant, R. (2006). Rapid Induction of Apoptosis in Gastrulating Mouse Embryos by Ethanol and its Prevention by HB-EGF. Alcohol Clin. Exp. Res. 30:127-134.
Loucks, E. and Ahlgrem, S. (2009). Deciphering the Role of SHH Signaling in Axial Defects Produced by Ethanol Exposure. Birth Defects Research. 85:556-567.
Marti, E., Takada, R., Bumcrot, D.A., Sasaki, H., McMahon, A.P. (1995). Distribution of Sonic Hedgehog Peptides in the Developing Chick and Mouse Embryo. Development. 121: 2537-2547.